Using `egor` to analyse ego-centered network data
Till Krenz
2025-10-31
Source:vignettes/using_egor.Rmd
using_egor.RmdThe egor Package
egor provides
- import functions
- egor object organizes ego-centered network, allowing for a smooth workflow
- dplyr methods: enable tidy data analysis strategies
- descriptive analysis (network composition, density, homophily, diversity)
- visualization (clustered graphs, egographs, egogram)
- interactive visualization app
An egor object contains all data levels associated with
ego-centered network analysis, those levels are: ego, alter, alter-alter
ties. By providing the egor()-function with
data.frames containing data corresponding to these data
levels, we construct an egor object. Here is an example of what the
data.frames could look like. Pay attention to the ID
variables connecting the levels with each other.
library(egor)| .ALTID | .EGOID | sex | age | age.years | country | income |
|---|---|---|---|---|---|---|
| 1 | 1 | m | 46 - 55 | 48 | USA | 45625 |
| 2 | 1 | m | 0 - 17 | 5 | Germany | 52925 |
| 3 | 1 | w | 26 - 35 | 35 | Australia | 60225 |
| 4 | 1 | w | 0 - 17 | 3 | Poland | 25550 |
| 5 | 1 | m | 66 - 100 | 97 | Australia | 45260 |
| 6 | 1 | w | 26 - 35 | 29 | Germany | 8395 |
| .EGOID | sex | age | age.years | country | income |
|---|---|---|---|---|---|
| 1 | m | 56 - 65 | 63 | Australia | 29930 |
| 2 | m | 26 - 35 | 33 | Germany | 17885 |
| 3 | m | 66 - 100 | 74 | Germany | 20805 |
| 4 | w | 18 - 25 | 21 | Poland | 29565 |
| 5 | m | 0 - 17 | 9 | Germany | 15330 |
| 6 | m | 0 - 17 | 6 | Australia | 23360 |
| .EGOID | .SRCID | .TGTID | weight |
|---|---|---|---|
| 20 | 1 | 2 | 0.6666667 |
| 25 | 6 | 10 | 0.6666667 |
| 9 | 6 | 8 | 0.6666667 |
| 31 | 2 | 10 | 0.6666667 |
| 24 | 1 | 12 | 0.3333333 |
| 11 | 9 | 11 | 0.3333333 |
All three data.frames contain an egoID identifying a
unique ego and connecting their personal data to the alter and
alter-alter tie data. The alterID is in the alter data is reused in the
alter-alter tie data in the Source and Target columns.
Let’s create an egor object from the data we just loaded.
e1 <- egor(alters = alters32,
egos = egos32,
aaties = aaties32,
ID.vars = list(
ego = ".EGOID",
alter = ".ALTID",
source = ".SRCID",
target = ".TGTID"))
e1
#> # EGO data (active): 32 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 m 56 - 65 63 Australia 29930
#> 2 2 m 26 - 35 33 Germany 17885
#> 3 3 m 66 - 100 74 Germany 20805
#> 4 4 w 18 - 25 21 Poland 29565
#> 5 5 m 0 - 17 9 Germany 15330
#> # ℹ 27 more rows
#> # ALTER data: 384 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 1 m 46 - 55 48 USA 45625
#> 2 2 1 m 0 - 17 5 Germany 52925
#> 3 3 1 w 26 - 35 35 Australia 60225
#> # ℹ 381 more rows
#> # AATIE data: 1,056 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 20 1 2 0.667
#> 2 25 6 10 0.667
#> 3 9 6 8 0.667
#> # ℹ 1,053 more rowsAn [egor] object is a [list] of three
[tibbles], named “ego”, “alter” and “aatie”, containing
ego, alter and alter-alter tie data.
Import
There are currently three importing functions that read the data from
disk and load them as an egor object.
read_openeddi()
read_egoweb()
read_egonet()In addition there are three functions that help with the transformation of common data formats of ego-centered network data into egor objects:
onefile_to_egor()
twofiles_to_egor()
threefiles_to_egor()Manipulate
Manipulating an egor object can be done with base R functions or with
dplyr verbs.
Base R
The different data levels of an egor object can be manipulated using
square bracket subsetting or the subset() function.
Ego level:
e1[e1$ego$age.years > 35, ]
#> # EGO data (active): 19 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 m 56 - 65 63 Australia 29930
#> 2 3 m 66 - 100 74 Germany 20805
#> 3 7 m 66 - 100 84 Australia 19345
#> 4 8 w 66 - 100 100 Poland 35040
#> 5 9 m 36 - 45 38 USA 64605
#> # ℹ 14 more rows
#> # ALTER data: 228 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 1 m 46 - 55 48 USA 45625
#> 2 2 1 m 0 - 17 5 Germany 52925
#> 3 3 1 w 26 - 35 35 Australia 60225
#> # ℹ 225 more rows
#> # AATIE data: 641 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 25 6 10 0.667
#> 2 9 6 8 0.667
#> 3 7 3 6 0.667
#> # ℹ 638 more rowsAlter level:
subset(e1, e1$alter$sex == "w", unit = "alter")
#> # EGO data (active): 32 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 m 56 - 65 63 Australia 29930
#> 2 2 m 26 - 35 33 Germany 17885
#> 3 3 m 66 - 100 74 Germany 20805
#> 4 4 w 18 - 25 21 Poland 29565
#> 5 5 m 0 - 17 9 Germany 15330
#> # ℹ 27 more rows
#> # ALTER data: 204 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 3 1 w 26 - 35 35 Australia 60225
#> 2 4 1 w 0 - 17 3 Poland 25550
#> 3 6 1 w 26 - 35 29 Germany 8395
#> # ℹ 201 more rows
#> # AATIE data: 300 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 25 6 10 0.667
#> 2 9 6 8 0.667
#> 3 7 3 6 0.667
#> # ℹ 297 more rowsAlter-alter tie level:
subset(e1, e1$aatie$weight > 0.5, unit = "aatie")
#> # EGO data (active): 32 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 m 56 - 65 63 Australia 29930
#> 2 2 m 26 - 35 33 Germany 17885
#> 3 3 m 66 - 100 74 Germany 20805
#> 4 4 w 18 - 25 21 Poland 29565
#> 5 5 m 0 - 17 9 Germany 15330
#> # ℹ 27 more rows
#> # ALTER data: 384 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 1 m 46 - 55 48 USA 45625
#> 2 2 1 m 0 - 17 5 Germany 52925
#> 3 3 1 w 26 - 35 35 Australia 60225
#> # ℹ 381 more rows
#> # AATIE data: 721 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 20 1 2 0.667
#> 2 25 6 10 0.667
#> 3 9 6 8 0.667
#> # ℹ 718 more rowsactivate() and dplyr verbs
An egor object can be manipulated with dplyr verbs.
Using the activate() command, the data level to execute manipulations
on, can be changed. This concept is borrowed from the tidygraph
package.
If the manipulation leads to the deletion of egos, the respective alters and alter-alter ties are deleted as well. Similarly deletions of alters lead to the exclusion of the alter-alter ties of the deleted alters.
e1 %>%
filter(income > 36000)
#> # EGO data (active): 10 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 9 m 36 - 45 38 USA 64605
#> 2 10 m 0 - 17 14 Australia 49275
#> 3 11 w 26 - 35 27 Germany 37960
#> 4 12 m 56 - 65 57 Germany 54750
#> 5 15 w 26 - 35 28 Germany 46720
#> # ℹ 5 more rows
#> # ALTER data: 120 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 9 m 46 - 55 48 USA 45625
#> 2 2 9 m 0 - 17 5 Germany 52925
#> 3 3 9 w 26 - 35 35 Australia 60225
#> # ℹ 117 more rows
#> # AATIE data: 333 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 20 1 2 0.667
#> 2 9 6 8 0.667
#> 3 11 9 11 0.333
#> # ℹ 330 more rows
e1 %>%
activate(alter) %>%
filter(country %in% c("USA", "Poland"))
#> # EGO data: 32 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 m 56 - 65 63 Australia 29930
#> 2 2 m 26 - 35 33 Germany 17885
#> 3 3 m 66 - 100 74 Germany 20805
#> # ℹ 29 more rows
#> # ALTER data (active): 180 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 1 m 46 - 55 48 USA 45625
#> 2 4 1 w 0 - 17 3 Poland 25550
#> 3 7 1 m 26 - 35 32 USA 54020
#> 4 8 1 w 46 - 55 49 USA 60955
#> 5 11 1 w 46 - 55 54 Poland 9490
#> # ℹ 175 more rows
#> # AATIE data: 218 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 31 2 10 0.667
#> 2 24 1 12 0.333
#> 3 7 3 6 0.667
#> # ℹ 215 more rows
e1 %>%
activate(aatie) %>%
filter(weight > 0.7)
#> # EGO data: 32 × 6
#> .egoID sex age age.years country income
#> * <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 m 56 - 65 63 Australia 29930
#> 2 2 m 26 - 35 33 Germany 17885
#> 3 3 m 66 - 100 74 Germany 20805
#> # ℹ 29 more rows
#> # ALTER data: 384 × 7
#> .altID .egoID sex age age.years country income
#> * <int> <dbl> <chr> <fct> <int> <chr> <dbl>
#> 1 1 1 m 46 - 55 48 USA 45625
#> 2 2 1 m 0 - 17 5 Germany 52925
#> 3 3 1 w 26 - 35 35 Australia 60225
#> # ℹ 381 more rows
#> # AATIE data (active): 374 × 4
#> .egoID .srcID .tgtID weight
#> * <int> <int> <int> <dbl>
#> 1 26 2 10 1
#> 2 15 6 10 1
#> 3 16 3 6 1
#> 4 24 2 8 1
#> 5 26 6 11 1
#> # ℹ 369 more rowsAnalyse
Try these function to analyse you egor object.
Summary
summary(e1)
#> 32 Egos/ Ego Networks
#> 384 Alters
#> Min. Netsize 12
#> Average Netsize 12
#> Max. Netsize 12
#> Average Density 0.5
#> Alter survey design:
#> Maximum nominations: InfDensity
ego_density(e1)
#> # A tibble: 32 × 2
#> .egoID density
#> <dbl> <dbl>
#> 1 1 0.485
#> 2 2 0.5
#> 3 3 0.5
#> 4 4 0.409
#> 5 5 0.561
#> 6 6 0.455
#> 7 7 0.652
#> 8 8 0.485
#> 9 9 0.515
#> 10 10 0.515
#> # ℹ 22 more rowsComposition
composition(e1, "age") %>%
head() %>%
kable()| .egoID | 0 - 17 | 18 - 25 | 26 - 35 | 36 - 45 | 46 - 55 | 56 - 65 | 66 - 100 |
|---|---|---|---|---|---|---|---|
| 1 | 0.1666667 | NA | 0.2500000 | NA | 0.3333333 | 0.0833333 | 0.1666667 |
| 2 | 0.3333333 | 0.1666667 | NA | 0.0833333 | 0.1666667 | NA | 0.2500000 |
| 3 | 0.1666667 | 0.1666667 | 0.0833333 | NA | 0.1666667 | 0.0833333 | 0.3333333 |
| 4 | 0.0833333 | 0.0833333 | 0.1666667 | NA | 0.2500000 | 0.0833333 | 0.3333333 |
| 5 | 0.2500000 | 0.1666667 | NA | 0.0833333 | 0.1666667 | NA | 0.3333333 |
| 6 | 0.1666667 | 0.0833333 | 0.2500000 | NA | 0.2500000 | 0.0833333 | 0.1666667 |
Diversity
alts_diversity_count(e1, "age")
#> # A tibble: 32 × 2
#> .egoID diversity
#> <dbl> <dbl>
#> 1 1 5
#> 2 2 5
#> 3 3 6
#> 4 4 6
#> 5 5 5
#> 6 6 6
#> 7 7 5
#> 8 8 5
#> 9 9 5
#> 10 10 5
#> # ℹ 22 more rows
alts_diversity_entropy(e1, "age")
#> # A tibble: 32 × 2
#> .egoID entropy
#> <dbl> <dbl>
#> 1 1 2.19
#> 2 2 2.19
#> 3 3 2.42
#> 4 4 2.36
#> 5 5 2.19
#> 6 6 2.46
#> 7 7 2.08
#> 8 8 2.13
#> 9 9 2.19
#> 10 10 2.19
#> # ℹ 22 more rowsEgo-Alter Homophily (EI-Index)
comp_ei(e1, "age", "age")
#> # A tibble: 32 × 2
#> .egoID ei
#> <dbl> <dbl>
#> 1 1 0.833
#> 2 2 1
#> 3 3 0.333
#> 4 4 0.833
#> 5 5 0.5
#> 6 6 0.667
#> 7 7 0.333
#> 8 8 0.333
#> 9 9 1
#> 10 10 0.333
#> # ℹ 22 more rowsEI-Index for Alter-Alter Ties
| .egoID | ei | 0 - 17 | 26 - 35 | 46 - 55 | 56 - 65 | 66 - 100 | 18 - 25 | 36 - 45 |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.5000000 | -0.1764706 | 0.25 | 1.0000000 | NaN | 1.0000000 | NA | NA |
| 2 | -0.0526316 | 0.1688312 | NA | -0.2500000 | NA | -0.3500000 | 1.0000000 | NaN |
| 3 | -0.1692308 | 1.0000000 | NaN | 1.0000000 | NaN | -0.2213740 | -0.2903226 | NA |
| 4 | 0.0132159 | NaN | 1.00 | -0.2413793 | NaN | 0.2000000 | NaN | NA |
| 5 | 0.0163934 | 1.0000000 | NA | -0.3333333 | NA | -0.0322581 | -0.3793103 | NaN |
| 6 | 0.1076923 | -0.3333333 | 1.00 | 0.1818182 | NaN | -0.3793103 | NaN | NA |
Count attribute combinations in alter-alter ties/ dyads
# return results as "wide" tibble
count_dyads(
object = e1,
alter_var_name = "country"
)
#> # A tibble: 32 × 11
#> .egoID dy_cou_Australia_Austr…¹ dy_cou_Australia_Ger…² dy_cou_Australia_Pol…³
#> <dbl> <int> <int> <int>
#> 1 1 2 6 3
#> 2 2 0 2 0
#> 3 3 4 6 4
#> 4 4 1 1 1
#> 5 5 2 11 4
#> 6 6 2 1 1
#> 7 7 0 5 7
#> 8 8 1 7 1
#> 9 9 1 6 4
#> 10 10 0 3 1
#> # ℹ 22 more rows
#> # ℹ abbreviated names: ¹dy_cou_Australia_Australia, ²dy_cou_Australia_Germany,
#> # ³dy_cou_Australia_Poland
#> # ℹ 7 more variables: dy_cou_Australia_USA <int>, dy_cou_Germany_Germany <int>,
#> # dy_cou_Germany_Poland <int>, dy_cou_Germany_USA <int>,
#> # dy_cou_Poland_USA <int>, dy_cou_USA_USA <int>, dy_cou_Poland_Poland <int>
# return results as "long" tibble
count_dyads(
object = e1,
alter_var_name = "country",
return_as = "long"
)
#> # A tibble: 278 × 3
#> .egoID dyads n
#> <dbl> <chr> <int>
#> 1 1 Australia_Australia 2
#> 2 1 Australia_Germany 6
#> 3 1 Australia_Poland 3
#> 4 1 Australia_USA 3
#> 5 1 Germany_Germany 3
#> 6 1 Germany_Poland 4
#> 7 1 Germany_USA 6
#> 8 1 Poland_USA 2
#> 9 1 USA_USA 3
#> 10 2 Australia_Germany 2
#> # ℹ 268 more rows
comp_ply()
comp_ply() applies a user-defined function on an alter
attribute and returns a numeric vector with the results. It can be used
to apply base R functions like sd(), mean() or
functions from other packages.
e2 <- make_egor(15, 32)
comp_ply(e2, "age.years", sd, na.rm = TRUE)
#> # A tibble: 15 × 2
#> .egoID result
#> <dbl> <dbl>
#> 1 1 26.5
#> 2 2 28.0
#> 3 3 25.5
#> 4 4 23.3
#> 5 5 25.0
#> 6 6 27.6
#> 7 7 27.7
#> 8 8 23.5
#> 9 9 26.5
#> 10 10 26.9
#> 11 11 24.8
#> 12 12 24.8
#> 13 13 26.5
#> 14 14 27.5
#> 15 15 27.8Visualize
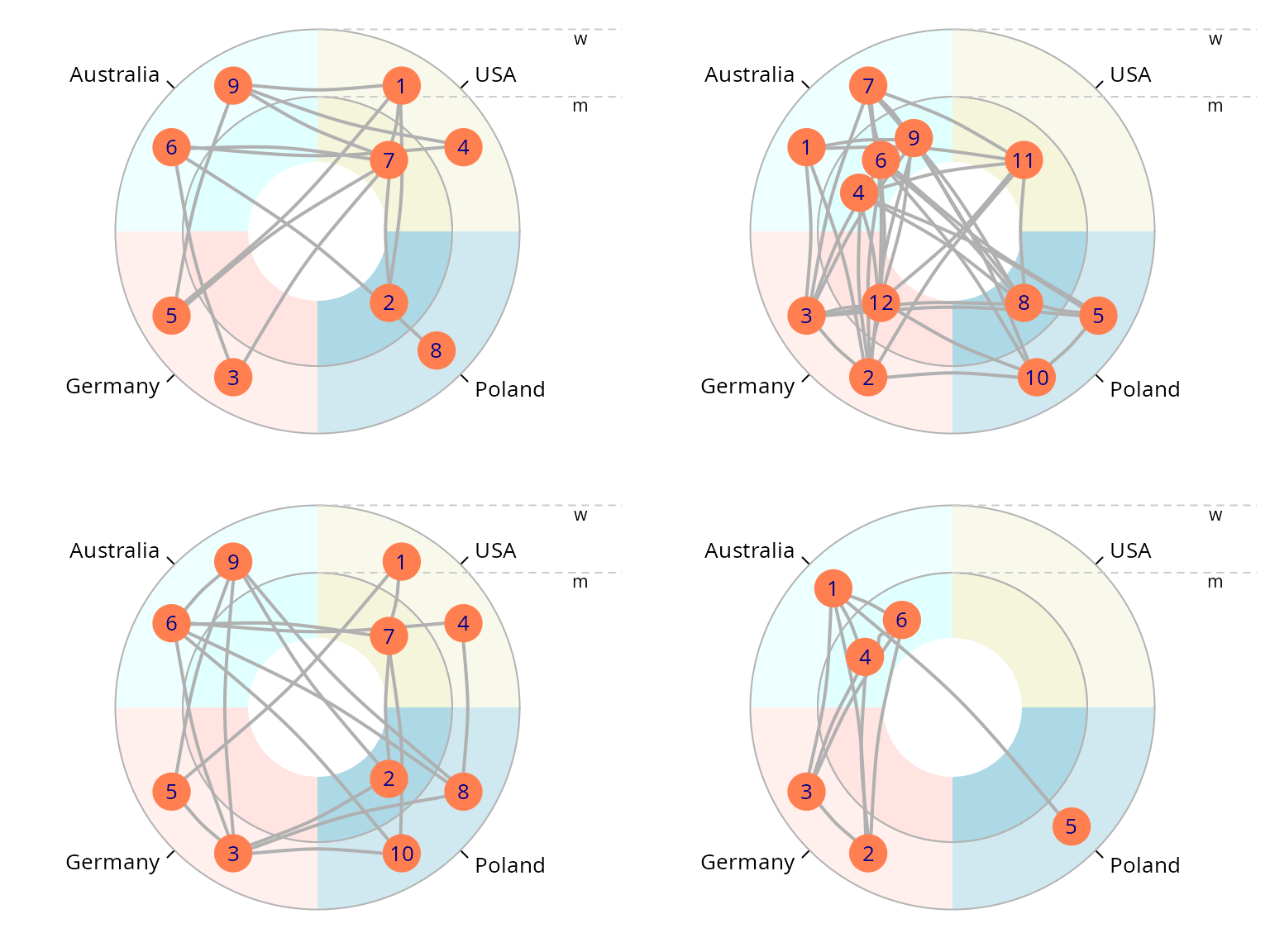
Clustered Graphs
data("egor32")
# Simplify networks to clustered graphs, stored as igraph objects
graphs <- clustered_graphs(egor32, "age")
# Visualize
par(mfrow = c(2,2), mar = c(0,0,0,0))
vis_clustered_graphs(graphs[1:3],
node.size.multiplier = 1,
edge.width.multiplier = 1,
label.size = 0.6)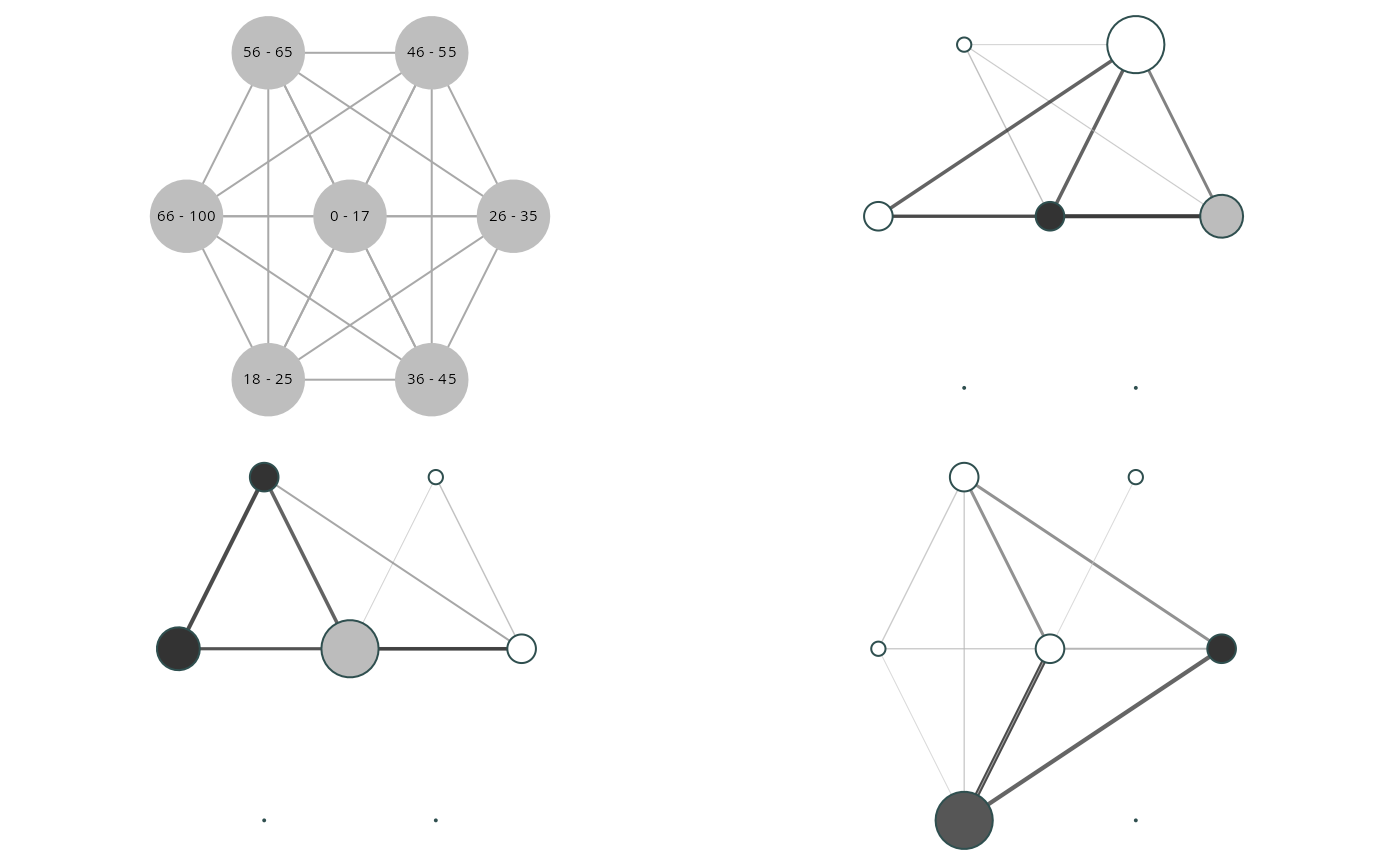
graphs2 <- clustered_graphs(make_egor(50, 50)[1:4], "country")
vis_clustered_graphs(graphs2[1:3],
node.size.multiplier = 1,
edge.width.multiplier = 3,
label.size = 0.6,
labels = FALSE)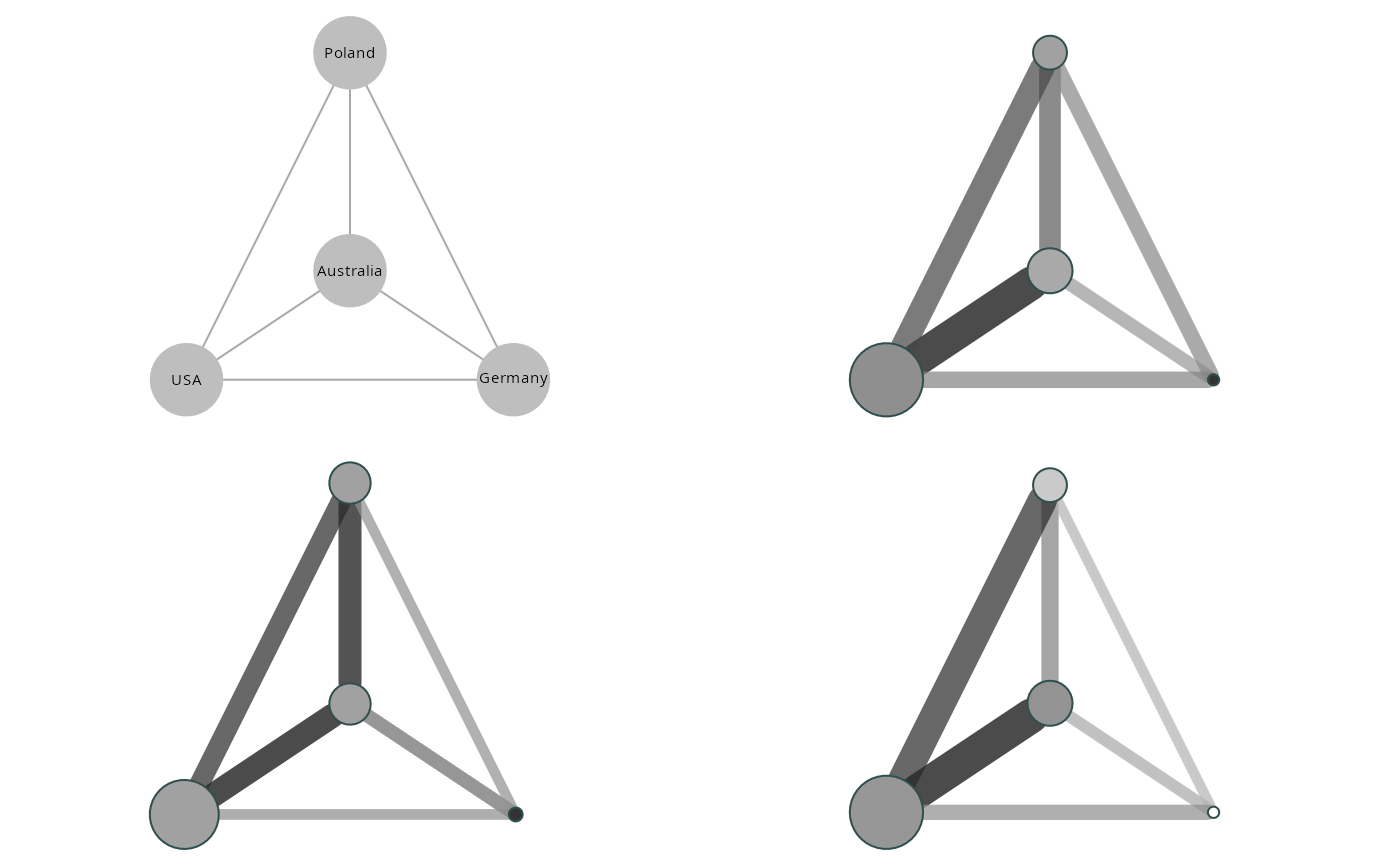
igraph & network plotting
-
as_igraph()converts anegorobject to a list of igraph objects. -
as_network()converts anegorobject to a list of network objects.
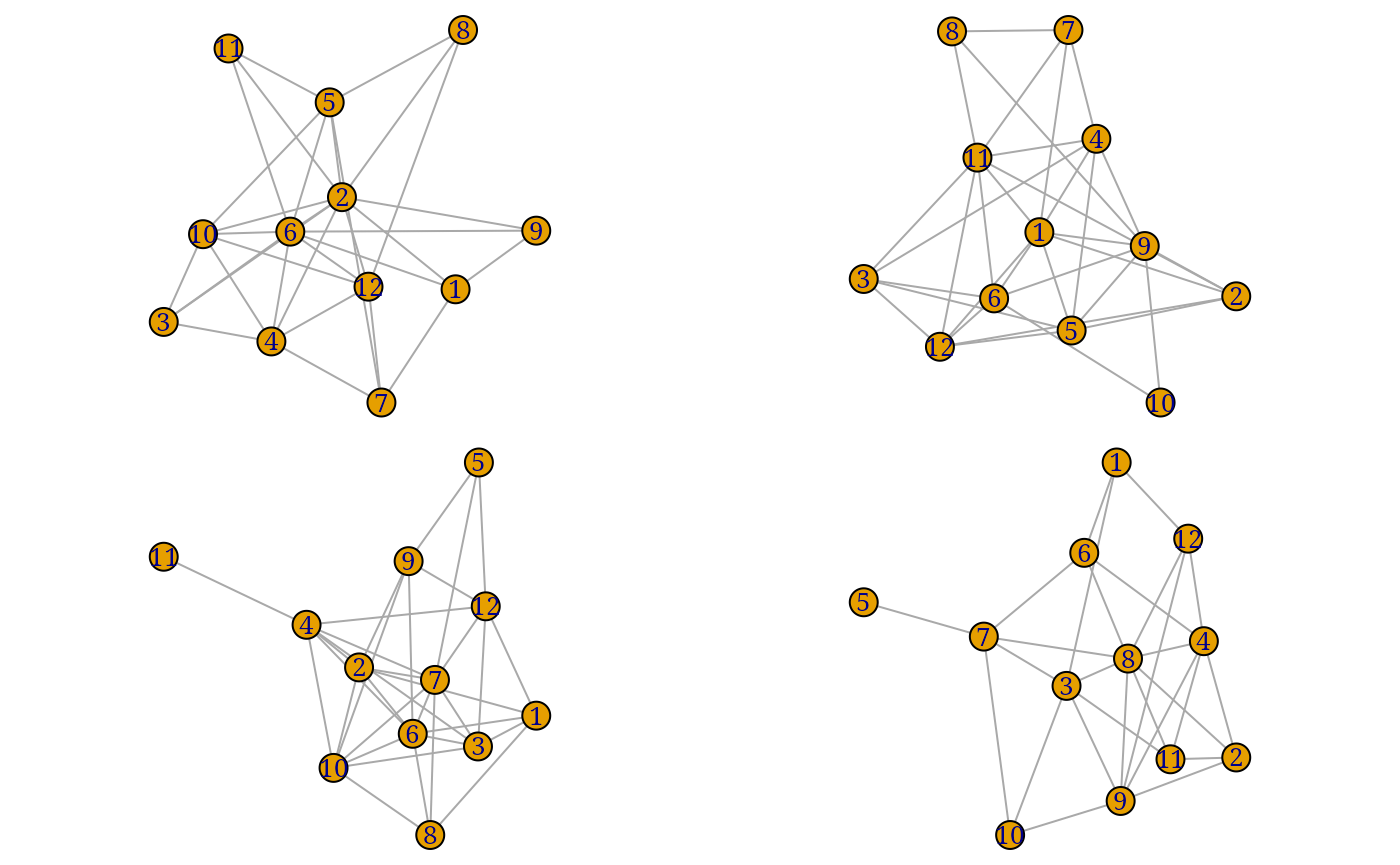
purrr::walk(as_network(egor32)[1:4], plot)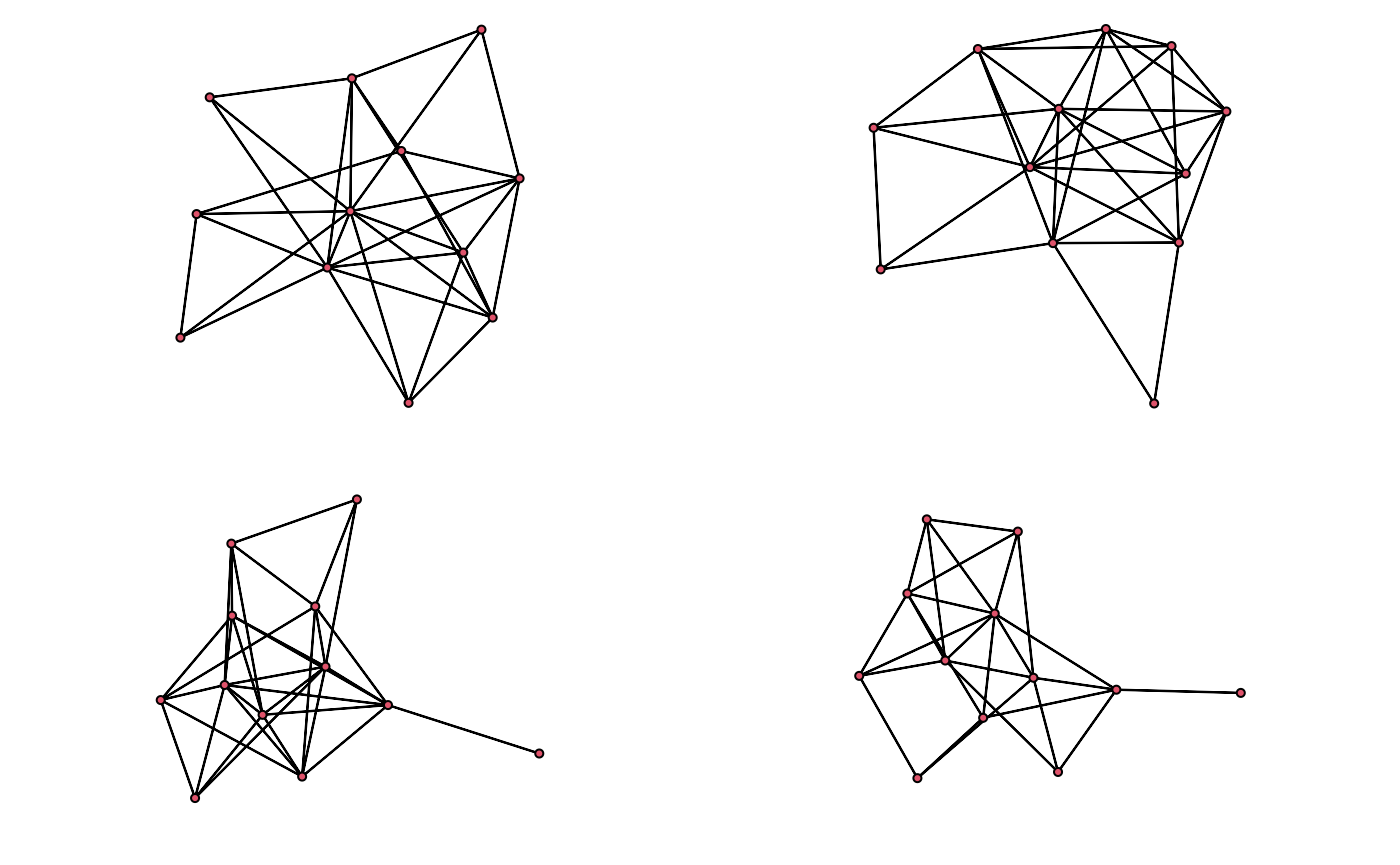
plot(egor32)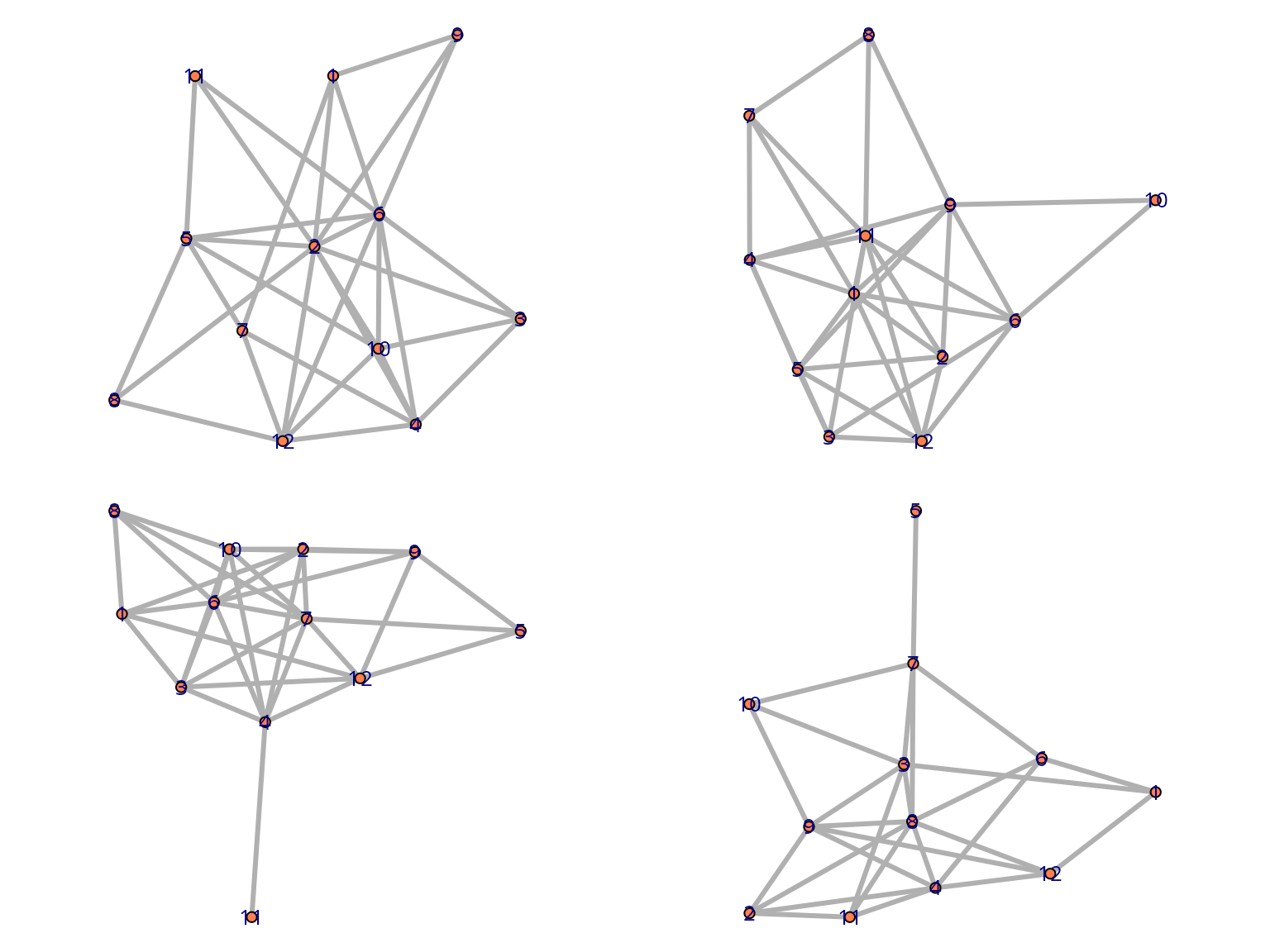
Shiny App for Visualization
egor_vis_app() starts a Shiny app which offers a
graphical interface for adjusting the visualization parameters of the
networks stored in an egor object.
egor_vis_app(egor32)Conversions
With as_igraph() and as_network() all ego
networks are transformed into a list of igraph/network objects.